The polymerase chain reaction
Overview
- Definition
- PCR simply explained
- PCR test simply explained
- PCR marker definition
- The procedure of a PCR
- A selection of our DNA polymerases
- A selection of our PCR master mixes
- Use of a PCR test to detect diseases
- HIV-PCR Test
- Reverse transcriptase polymerase chain reaction
- A selection of our products for RT-PCR
- Real Time Quantitative PCR
- A selection of our products for Real Time Quantitative PCR
- Current: Real Time Quantitative PCR for the detection of SARS-CoV-2
- Current: A selection of our products for the detection of SARS-CoV-2 with RT-qPCR
- Possible PCR error sources
- Further considerations
- Frequently asked questions
Definition
PCR is an abbreviation for Polymerase Chain Reaction (engl.).
The aim of such a polymerase chain reaction is to amplify the DNA, i.e. genetic material. This is necessary, for example, if there is not enough sample DNA available.
PCR simply explained
If the name is broken down into its constituent parts, the underlying principle of this procedure can be explained:
An enzyme called "DNA polymerase" is used within a biochemical reaction. In addition, it is a chain reaction: the end product of the reaction also serves as the starting point for a further, similar reaction.
PCR test simply explained
A PCR test is a method to check for the presence of a certain starting material.
By amplifying the starting material using PCR, it can be subsequently detected.
For example, a COVID-19 PCR test amplifies the viral genetic material. Even if this is only found in small amounts in the sample from mucous membranes of the respiratory tract, PCR multiplies it to such an extent that it can be detected.
Even a hair, as often seen in crime films, is sufficient to multiply the genetic material it contains to such an extent that a genetic profile of the perpetrator can be established.
PCR marker definition
PCR markers are like a ruler that serve to classify the size and mass of the PCR product. For example, a plasmid can be used that is digested using a restriction enzyme. The size and mass of the digested fragments are known and thus help to estimate the mass of the amplified PCR product during gel electrophoresis.
These PCR markers can be used to identify the size and mass of the PCR product.
These PCR markers are comparable to a protein ladder in an SDS-PAGE.
The procedure of a PCR
How does a PCR work?
The PCR can be divided into three sections: Denaturation, primer hybridization and elongation.
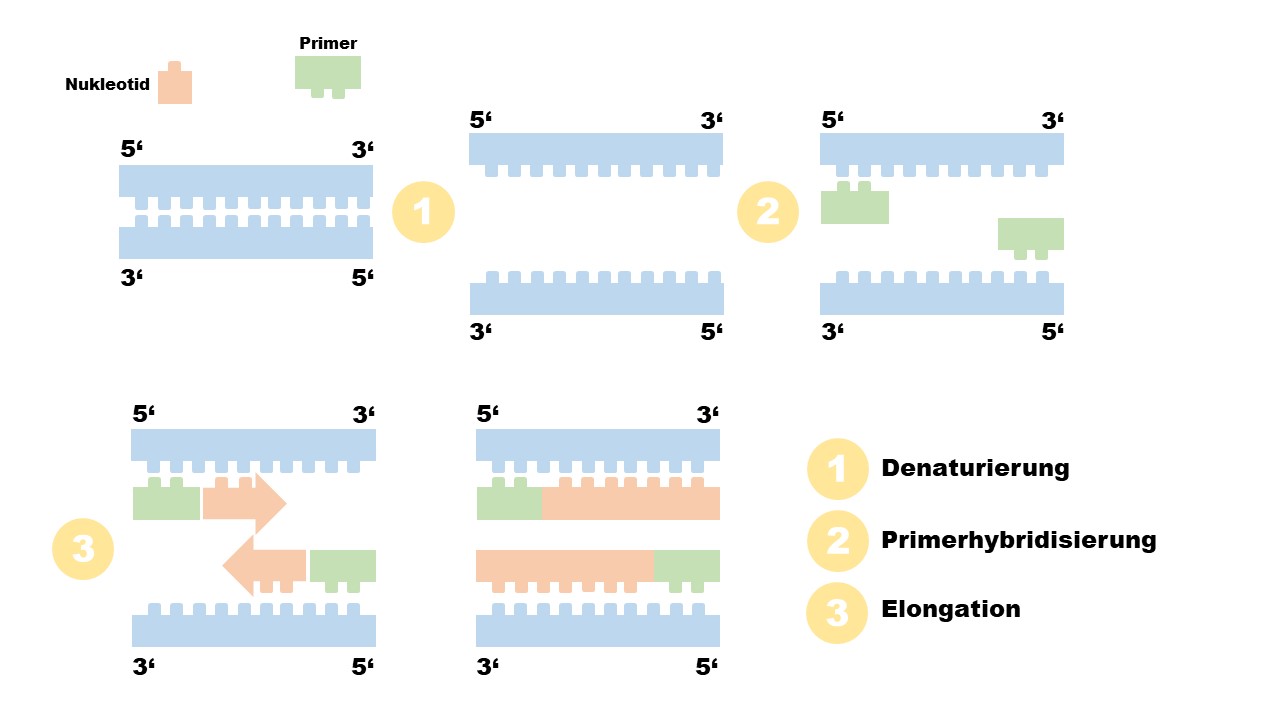 Img.1.: A polymerase chain reaction is divided into three characteristic steps. During denaturation the two DNA strands are separated. This is followed by primer hybridization in which the primers bind to their respective starting matrices. Finally, the synthesis of the "new material" takes place during the elongation .
Img.1.: A polymerase chain reaction is divided into three characteristic steps. During denaturation the two DNA strands are separated. This is followed by primer hybridization in which the primers bind to their respective starting matrices. Finally, the synthesis of the "new material" takes place during the elongation .
In the first step, denaturation, the two single strands of the double-stranded DNA are separated from each other with the help of heat. This breaks the hydrogen bonds that previously held both strands together.
For the next step, the primer hybridization, primers are necessary. Primers are oligonucleotides. They were selected to bind (hybridize) to a specific segment within the strand to be amplified.
The PCR primer design plays a special role in the course of a PCR. Various criteria must be taken into account to ensure error-free binding to the starting DNA. It is therefore not uncommon for several variants to be created and tested in practice.
In the next step, the polymerase can then bind to these primers and start replicating the DNA.
The process of actual amplification is called extension, elongation or amplification. Here, DNA polymerase migrates the nucleotides of the DNA single strand piece by piece. Meanwhile, it creates a complementary counterpart to the original strand with the help of free nucleotides (deoxy nucleotides = dATP, dGTP, dCTP, dTTP). This is therefore an identical copy of the other single strand.
Each of these processes has a particular temperature. This is the reason why a thermostable DNA polymerase is used like for example the Pfu- or Tag-Polymerase. Furthermore, a constant change depending on the reaction step and cycle is not possible manually, therefore a PCR is performed in a so-called thermal cycler. As the name already describes, this is a PCR machine that runs through different temperatures in defined cycles.
A selection of our DNA polymerases
| Product | Supplier | Art. No. | Amount | |
|---|---|---|---|---|
| TAQ DNA POLYMERASE |
1000.0U |
|||
| RECOMBINANT TAQ DNA POLYMERASE |
1,000U |
|||
| EXPRIME TAQ DNA POLYMERASE (WITH DNTP MIXTURE AND 10X RXN BUFFER (WITH MG)) |
1,000U |
|||
| HS PRIME TAQ DNA POLYMERASE (WITH 10MM DNTP MIX., 25MM MGCL2 & 10X RXN BUFFER(MG FREE)) |
250 Units |
|||
| TAQ DNA POLYMERASE (RECOMBINANT) |
1,000U |
|||
| TAQ DNA POLYMERASE, 5U/UL |
BB-B0089-200 |
200U |
||
| FAST-TAQ DNA POLYMERASE WITH 10MM DNTP MIX |
500U |
|||
| HIGH-TAQ DNA POLYMERASE WITH 10MM DNTP MIX |
250U |
|||
| AT TAQ DNA POLYMERASE (HOTSTART - 5U/UL) |
200U |
|||
| HOTSTART TAQ DNA POLYMERASE (5U/UL) |
250U |
To make the procedure of a PCR as simple as possible a PCR Mastermix is often used. Such a PCR mastermix contains the DNA polymerase, a suitable reaction buffer, as well as the required nucleotides, which serve as building blocks for the new "copies".
A selection of our PCR master mixes
| Product | Supplier | Art. No. | Amount | |
|---|---|---|---|---|
| EZWAY MULTIPLEX PCR MASTERMIX (2X) |
1 ml |
|||
| EZWAY MULTIPLEX PCR QMASTERMIX (2X) |
1 ml |
|||
| 2X HOTSTART TAG PCR MASTERMIX WITH LOADING DYE |
1 ml |
|||
| EXPRESS HI FI DNA POL, 2X MASTERMIX |
2,5 ml (100 Reactions) |
|||
| VIPRIMEPLUS ONESTEP QRT-PCR MASTERMIX |
1ea |
Use of a PCR test to detect diseases
The PCR method can be advantageous not only for the amplification of DNA for an upcoming experiment.
Hereditary diseases are caused by changes in the genome. The specific gene can be amplified by means of PCR. Subsequently, the material is sequenced to detect possible mutations.
A PCR test can also help in the diagnosis of viral diseases such as an infection with the HI virus. In this test the viral DNA is amplified and can then be examined. In the case of RNA viruses, the RNA is transcribed into cDNA by a reverse transcriptase. This allows amplification by PCR as described above (RT-PCR).
HIV-PCR Test
A PCR test can also be used for the detection of HIV. It offers a central difference to other tests: While other tests detect antibodies, an HIV PCR test detects the HIV virus.
However, it should be emphasized that this HIV-PCR test is more likely to be used in the context of therapy or in connection with an antibody screening test than as a control test. Here, the previously obtained result should only be confirmed again.
Reverse transcriptase polymerase chain reaction
Similar to PCR, the reverse transcriptase polymerase chain reaction (RT-PCR) is divided into three steps: Purification of the RNA, reverse transcription of the RNA into DNA and amplification of the cDNA thus generated.
While we have previously learned about polymerase, this variant of PCR is followed by the introduction of another enzyme: reverse transcriptase. This enzyme originally comes from retroviruses. However, this enzyme is further modified for industrial use. For example, its RNase H activity has usually been removed.
The reverse transcriptase is also a DNA polymerase. However, the difference to the "normal" polymerase is that the reverse transcriptase requires RNA as starting material. It is therefore also known as RNA-dependent DNA polymerase. Using a DNA primer, the reverse transcriptase is able to produce complementary DNA (cDNA).
The difference between DNA and cDNA is that the latter has no introns. This is due to the fact that cDNA is produced by means of an RNA strand that has already gone through the splicing process.
A selection of our products for RT-PCR
| Products | Supplier | Art. No. | Amount | |
|---|---|---|---|---|
| One-Step RT-PCR Kit |
5 Reactions |
|||
| One Step RT PCR Kit (cDNA Synthesis + PCR In One Reaction) |
100 Reactions |
Real Time Quantitative PCR
Real Time Quantitative PCR (qPCR for short) is a common PCR in which the synthesized DNA is additionally quantified. The term "real time" refers to the quantification process: the increase in material is measured in real time using fluorescence.
One possibility is, for example, the use of DNA dyes. Commonly used are ethidium bromide or SYBR Green I. After successful synthesis of the complementary strand, the starting material is again available as a double-helix strand. The corresponding dyes can then intercalate into the DNA and thus stain it. The increase in synthesized material is proportional to the fluorescence intensity of the reaction batch.
A selection of our products for Real Time Quantitative PCR
| Products | Supplier | Art. No. | Amount | |
|---|---|---|---|---|
| SYBR GREEN I NUCLEIC ACID GEL STAIN |
100 µg |
|||
| Prime Q-Master Mix (2X conc. with SYBR Green I) |
100 ml |
|||
| SUPRIMESCRIPT QRT-PCR KIT (FOR TAQMAN PROBE, 2X CONC., WITH ROX DYE) |
100 Reactions |
|||
| SYBR GREEN I NUCLEIC ACID GEL STAIN |
100 µl |
|||
| RT MASTER MIX FOR QPCR |
1 ml (100 Reactions) |
|||
| KOD SYBR QPCR MIX |
1 ml |
|||
| FAST EVAGREEN MASTER MIX FOR QPCR(200 RXN) |
1 ml |
|||
| CYBRFAST 1-STEP RT-QPCR HI-ROX KIT |
100 Reactions |
|||
| VALIDATED FLUORESCENT PROBES FOR REAL-TIME QUANTITATIVE PCR (TAQMAN PROBE: FAM-BHQ1) |
200 Reactions |
Real time quantitative PCR for the detection of SARS-CoV-2
In view of the rapid spread of SARS-CoV-2, the demand for direct and indirect detection methods for COVID-19 has increased rapidly worldwide. Indirect detection methods include serological tests based on either SARS-CoV-2 antibody detection or SARS-CoV-2 antigen detection. In most cases, however, such tests must be combined with direct virus detection. RT-qPCR is the most widely used test.
A selection of our products for the detection of SARS-CoV-2 by RT-qPCR
| Products | Supplier | Art. No. | Tests/RXNs |
|---|---|---|---|
|
SARS-CoV-2 qPCR detection 1-plex assay-ORF1ab SARS-CoV-2 qPCR detection 1-plex assay-N SARS-CoV-2 qPCR detection 1-plex assay-RdRP SARS-CoV-2 qPCR detection 1-plex assay-E |
100 rxns |
||
| ABScript II One Step RT-qPCR Probe Kit |
20 rxns |
||
| Coronavirus (SARS-CoV-2) Real Time RT-PCR Nucleic Acid Detection Kit |
20 Tests |
||
| COVID-19 RT-qPCR Rapid Detection Kit |
100 rxns |
Possible PCR error sources
There are many things that can (but need not) go wrong with a PCR.
For instance, the correct concentration of the individual reagents plays an important role in most cases. Special attention should be paid to the dNTPS, the MgCl solution, the polymerase and the template.
TIP: Especially as a newcomer to the laboratory it helps to exchange information with colleagues and ask for their standard PCR protocol. These are usually long established. If the opportunity arises, you might even be able to use their preset thermal cycler program.
An incorrect thermal cycler program can also be a possible source of PCR errors.
However, if the DNA polymerase does not start working at all, no material can be amplified in a PCR. This might be due to a faulty primer design, for example.
TIP: It is recommended to synthesize several primer pairs at the beginning of the experiment. So you always have a back-up if one pair does not work and if all of them bind you have an additional control.
Further considerations
Maximum length of a target DNA that can be amplified by PCR
The length of the target DNA that can be amplified by PCR is limited by factors such as the specificity of the primers and the efficiency of the Taq polymerase, so it is important to understand these limitations in order to choose the appropriate technique for their experiments.
Optimal annealing temperature for PCR amplification of a specific target gene
The decision for a specific annealing temperature helps determining the efficiency of the primer binding to its target DNA.
Thus, it affects the specificity and yield of the PCR reaction.
Frequently asked questions:
What is a PCR test?
A PCR test is a detection test in which the presence of a specific DNA (or RNA) is shown by its amplification.
What is PCR diagnostics?
In diagnostics, for example, a PCR test can be used to detect an underlying genetic defect.
How long does a PCR test take?
The duration of a PCRT test depends on the number of cycles used.
Who invented PCR?
Kary Mullis is the inventor of PCR.
What is a primer in biology?
A primer is an oligonucleotide. This single-stranded DNA fragment serves as a starting point for DNA polymerase and is specifically used within a PCR.
Which enzyme removes the primers?
The 5'-3' exonuclease activity of Polymerase I or RNase H removes primers in prokaryotes.
Why are there Okazaki fragments?
Due to the 5'-->3' synthesis activity of DNA polymerase, discontinuous small DNA fragments are generated on the subsequent strand (complementary to the leader strand). These sections are called Okazaki fragments.
What is the most commonly used Taq polymerase in PCR?
Taq polymerase is the most commonly used DNA polymerase in PCR.
What is the effect of Mg2+ concentration on PCR amplification efficiency?
Mg2+ ions are important cofactors for Taq polymerase, and varying their concentration can affect the yield and specificity of the PCR reaction.
What is the difference between hot-start and normal PCR?
Hot-start PCR is a modification of normal PCR that aims to reduce non-specific amplification by preventing the Taq polymerase from being active at the start of the reaction.
What is the ideal primer concentration?
PCR primer concentrations of 0.1 and 1.0 µM usually prove to be optimal.



